Publications
Recent Publications
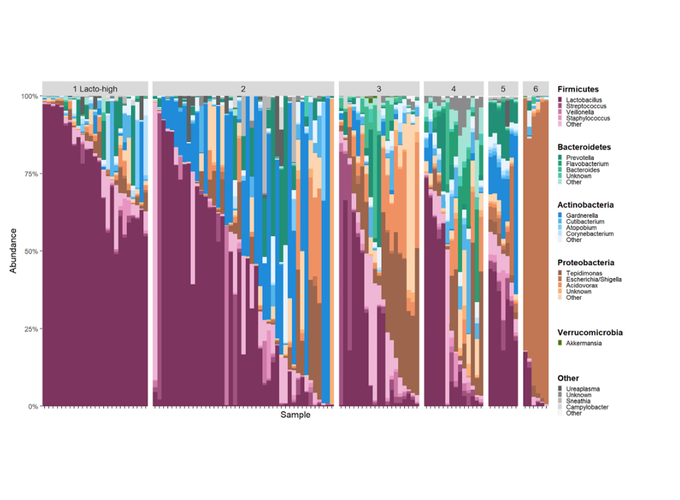
American Journal of Obstetrics & Gynecology
Urinary microbiome community types associated with urinary incontinence severity in women
October 2023
Abstract
This study aimed to test whether specific urinary or vaginal microbiome community types are associated with urinary incontinence severity in a population of women with mixed urinary incontinence
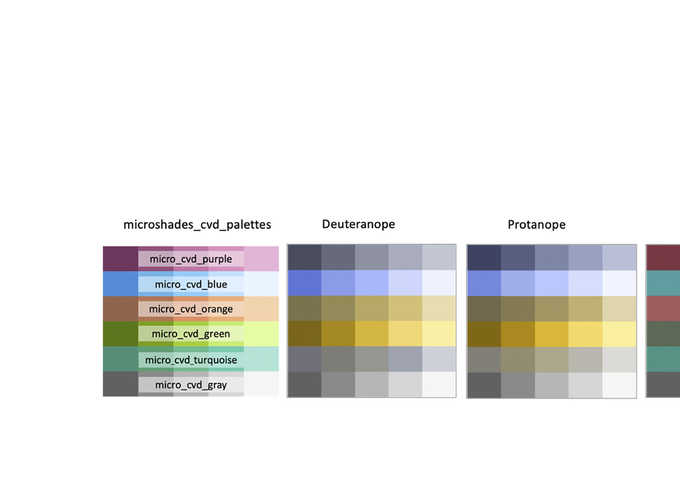
Microbiology Resource Announcements
microshades: An R Package for Improving Color Accessibility and Organization of Microbiome Data
October 2022
Abstract
When creating figures, it is important to consider that individuals with color vision deficiency (CVD) may not perceive all colors. While there are several CVD-friendly color palettes, they are often insufficient for working with microbiome data. Here, we introduce microshades, an R package for creating CVD-accessible microbiome figures.
DOI: 10.1128/mra.00795-22
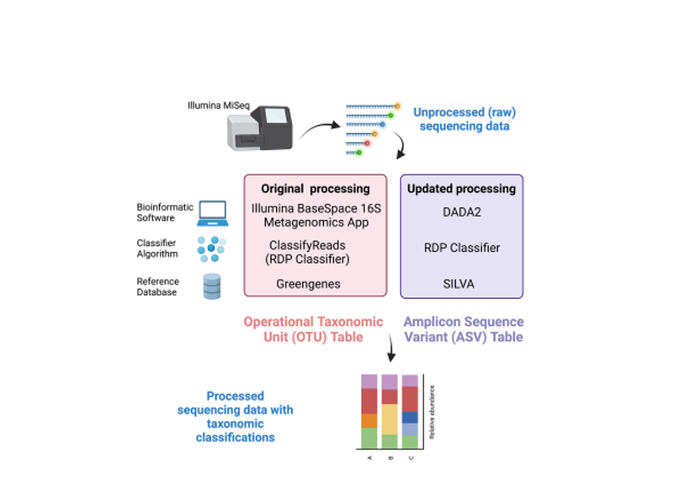
Frontiers
Updating Urinary Microbiome Analyses to Enhance Biologic Interpretation
July 2022
Abstract
An approach for assessing the urinary microbiome is 16S rRNA gene sequencing, where analysis methods are rapidly evolving. This re-analysis of an existing dataset aimed to determine whether updated bioinformatic and statistical techniques affect clinical inferences.

Female Pelvic Medicine & Reconstructive Surgery
The Impact of Local Estrogen on the Urogenital Microbiome in Genitourinary Syndrome of Menopause: A Randomized-Controlled Trial
June 2022
Abstract
The aims of this study were to characterize the effect of vaginal estrogen on the vaginal and urinary bladder microbiome in postmenopausal women and describe any clinical associations with the symptoms of genitourinary syndrome of menopause.
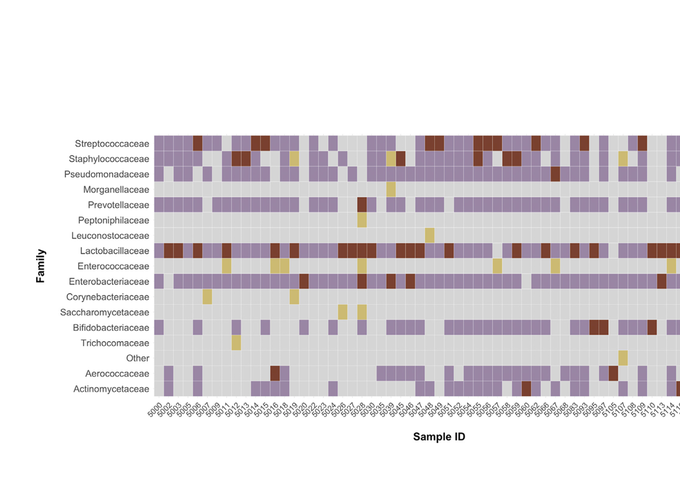
American Journal of Obstetrics & Gynecology
Concordance of urinary microbiota detected by 16S ribosomal RNA amplicon sequencing vs expanded quantitative urine culture
June 2022
Abstract
Multiple techniques exist for characterizing the urinary microbiome. Two commonly used methods include expanded quantitative urine culture (EQUC) and 16S ribosomal RNA amplicon sequencing (16S sequencing). Even with enhanced techniques, culture-based methods may still fail to detect several organisms that are difficult to grow.1,2 Sequencing methods also have limitations, such as failure to identify organisms with thick cell walls (eg, Gram-positive bacteria) owing to inefficient cytolysis.3 Given that each method of investigating the urobiome may present intrinsic biases toward detecting specific organisms, we aimed to compare the results between EQUC and 16S sequencing when both methods were performed on the same samples.
